Early Bird Trees Available for Download
Phylogenetic trees generated by the Early Bird project are available both as nexus format files and phylip (newick) format files. Branch length information is also provided in both file formats. Trees are available from:
- Hackett, Kimball, Reddy, and 15 additional co-authors (2008) Science, 320: 1763-1768. [list of files for download]
- An ultrametric version of the Hackett et al. (2008) tree (a timetree) that was used by Braun et al. (2011) and Han et al. (2011) is now available from the Hackett et al. (2008) trees. [link to ultrametric tree]
- Harshman, E. Braun, M. Braun, and 14 additional co-authors (2008) PNAS, 105: 13462-13467. [list of files for download]
- Galliform phylogenies from the Kimball-Braun lab group, including those from:
- Cox et al. 2007, Auk 124: 71-84.
- Kimball and Braun (2008) J Avian Biology, 39: 438-445.
- Bonilla et al. 2010, MPE 56: 536-542.
- Kimball et al. 2011, Int J Evol Biol Article ID 423938.
- Chojnowski et al. (2008) Gene, 410: 89-96.
Full information about these papers is available on the publications page. Trees generated as part of many of these studies have been deposited in TreeBase. The purpose of this page is to add value to the information available in public databases and to simplify further curation of the trees produced by the Early Bird group.
Trees from Hackett et al. 2008
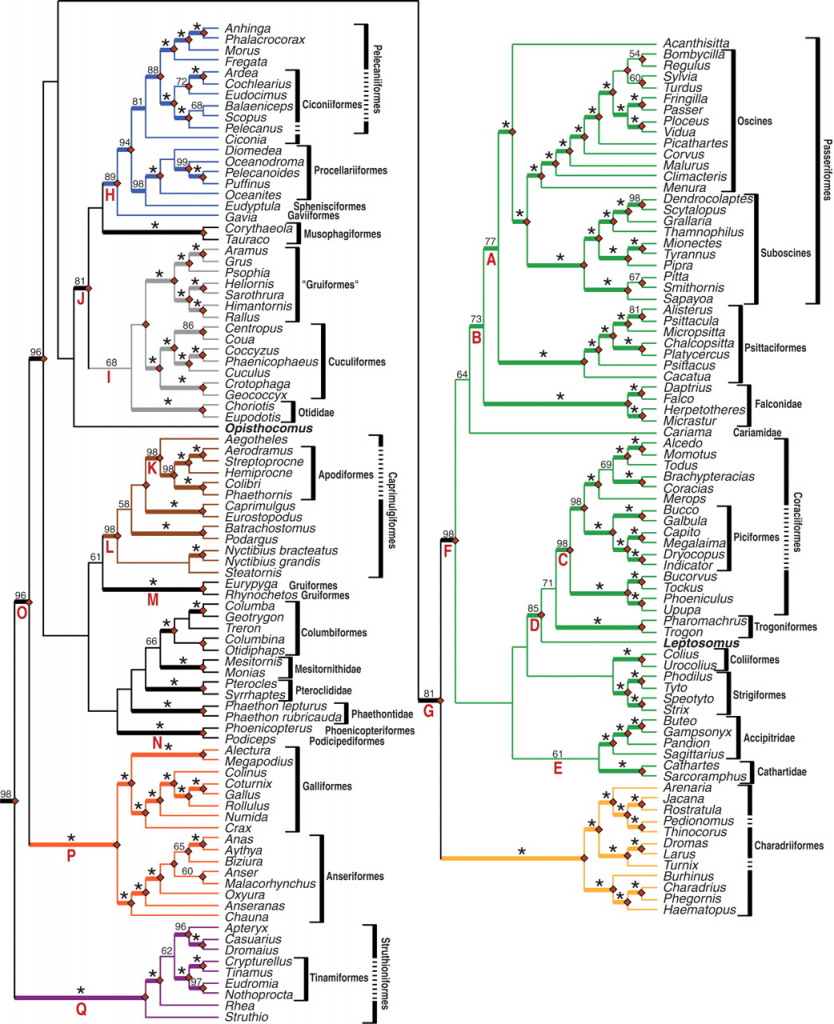
The Hackett et al. (2008) trees were based upon a total of 19 gene regions. Branch lengths were estimated by ML using the GTR+I+G model in PAUP* after excluding difficult to align regions in the dataset (download the paper and the alignments for additional details).
The Hackett et al. (2008) analyses revealed two slightly different trees. One topology is the ML bootstrap consensus (Fig. 2; also shown below) and the other is the optimal (ML) topology (Fig. 3 in Hackett et al. 2008; also presented as Fig. S1 in the supporting information). The only difference between the two topologies is the position of the Bombycilla + Regulus clade (click on the image of Fig. 2 for more information).
- Trees from Hackett et al. (2008) have been deposited in TreeBase with study ID S2108 (previously identified under the legacy study ID S2079)
- Cladograms with full taxon names and outgroups in nexus format.
- Trees with ML branch length estimates are included below. The nexus format treefiles include GTR+I+G model parameter estimates obtained using PAUP*.
- Rooted trees (include crocodilian outgroups) with ML branch length estimates. Includes both the ML bootstrap consensus tree (Fig. 2) and the ML tree (Fig. 3). [nexus format (both trees)] [phylip format (Figure 2)] [phylip format (Figure 3)]
- Identical trees without crocodilian outgroups. [nexus format (both trees)] [phylip format (Figure 2)] [phylip format (Figure 3)]
- All of these treefiles can be downloaded. The download includes two perl scripts that simplify renaming taxa in trees.
- An ultrametric version of the Fig. 3 topology from Hackett et al. (2008) generated by non-parametric rate smoothing. This timetree was used in Braun et al. (2011) and Han et al. (2011). [nexus format]
Trees from Harshman et al. 2008
A total of 20 gene regions were used to examine relationships among paleognaths (ratites and tinamous) in Harshman et al. (2008):
- Trees from Hackett et al. (2008) have been deposited in TreeBase with study ID S2134 (previously identified under the legacy study ID S2138)
- Analyses of the 20 gene supermatrix. [COMING SOON]
- Alignments of individual gene regions. [ALDOB] [BDNF] [CLTC] [CLTCL1] [CRYAA] [EEF2] [EGR1] [FGB] [IRF2] [GH1] [HMGN2] [MB] [MUSK] [MYC] [NGF] [NTF3] [PCBD1] [RHO] [TGFB2] [TPM1] ALL LINKS ARE COMING SOON
Gamebird (order Galliformes) trees
A number of galliform trees based upon multiple nuclear loci have been generated by the Kimball-Braun lab group:
- Trees from Cox et al. (2007) [nexus format]
- Tree from Figure 1 in Kimball and Braun (2008). [nexus format]
- Trees from Kimball et al. (2011) [pdf of tree figure] [nexus format file of optimal trees] [nexus format file of bootstrap tree] [zipped tarball of trees from the Bayesian analysis]
Trees from Chojnowski et al. 2008
Phylogenetic trees generated by analyzing two parlogous clathrin heavy chain genes (CLTC and CLTCL1):
- ML tree based for the combined (two gene) clathrin heavy chain matrix. [nexus format]
Molecular clock analyses were presented in the supplementary material to Chojnowski et al. (2008) along with information about the taxa that were examined. [pdf format]